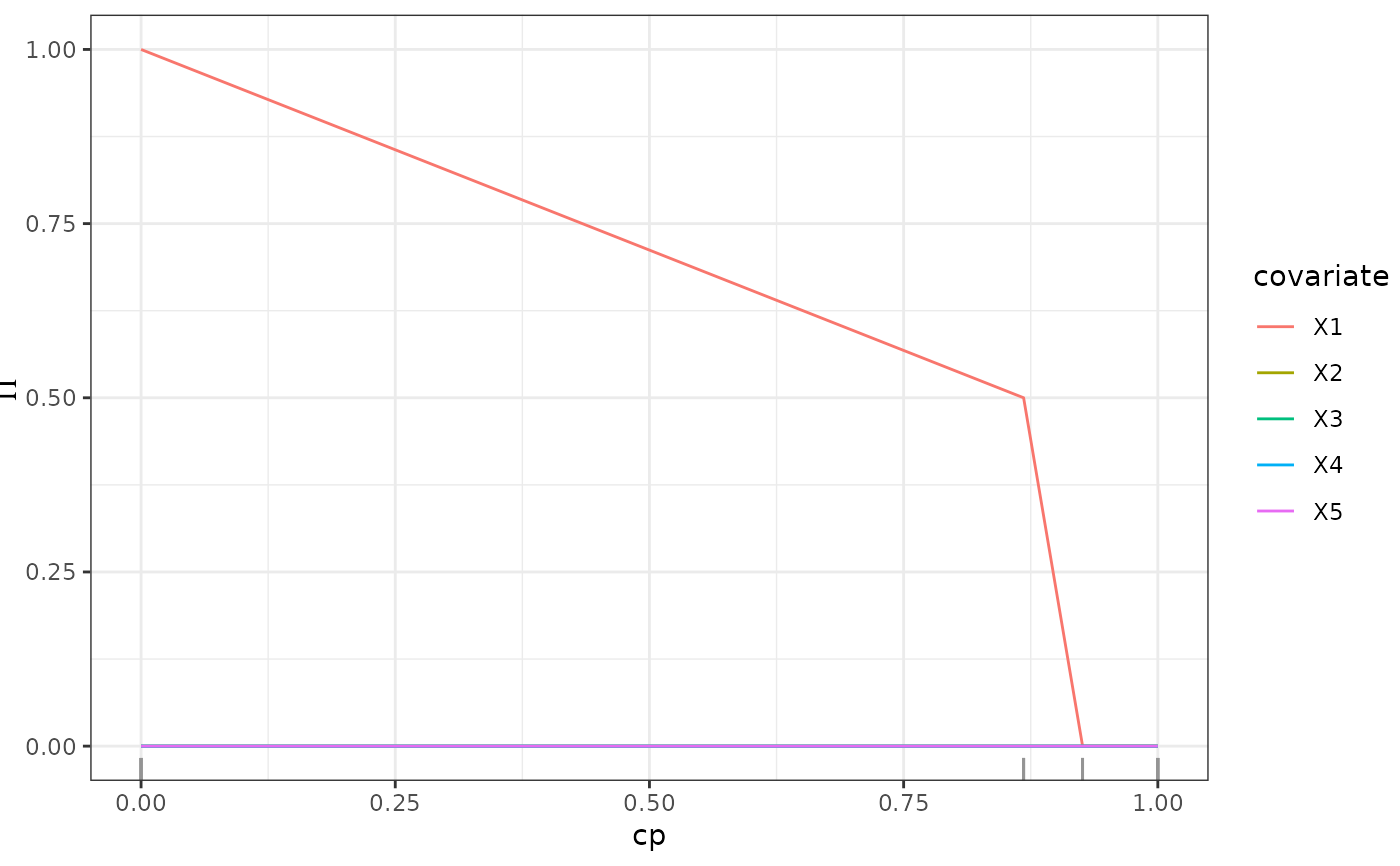
This function calculates the stability selection of an SDForest Meinshausen2010StabilitySelectionSDModels. Stability selection is calculated as the fraction of trees in the forest that select a variable for a split at each complexity parameter.
# S3 method for class 'SDForest'
stabilitySelection(object, cp_seq = NULL, ...)Arguments
Value
An object of class paths containing
- cp
The sequence of complexity parameters.
- varImp_path
A
matrixwith the stability selection for each complexity parameter.- type
Path type