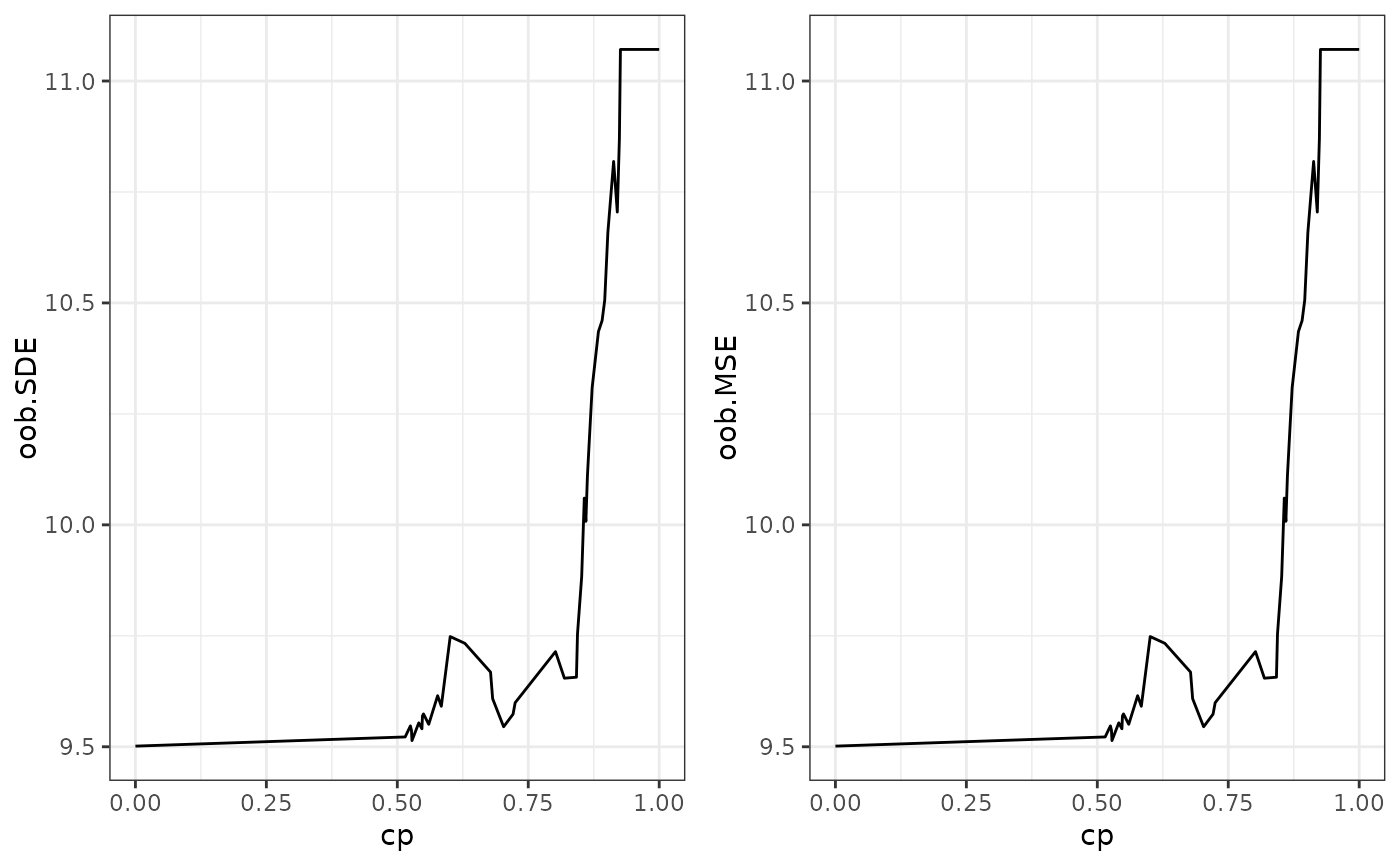
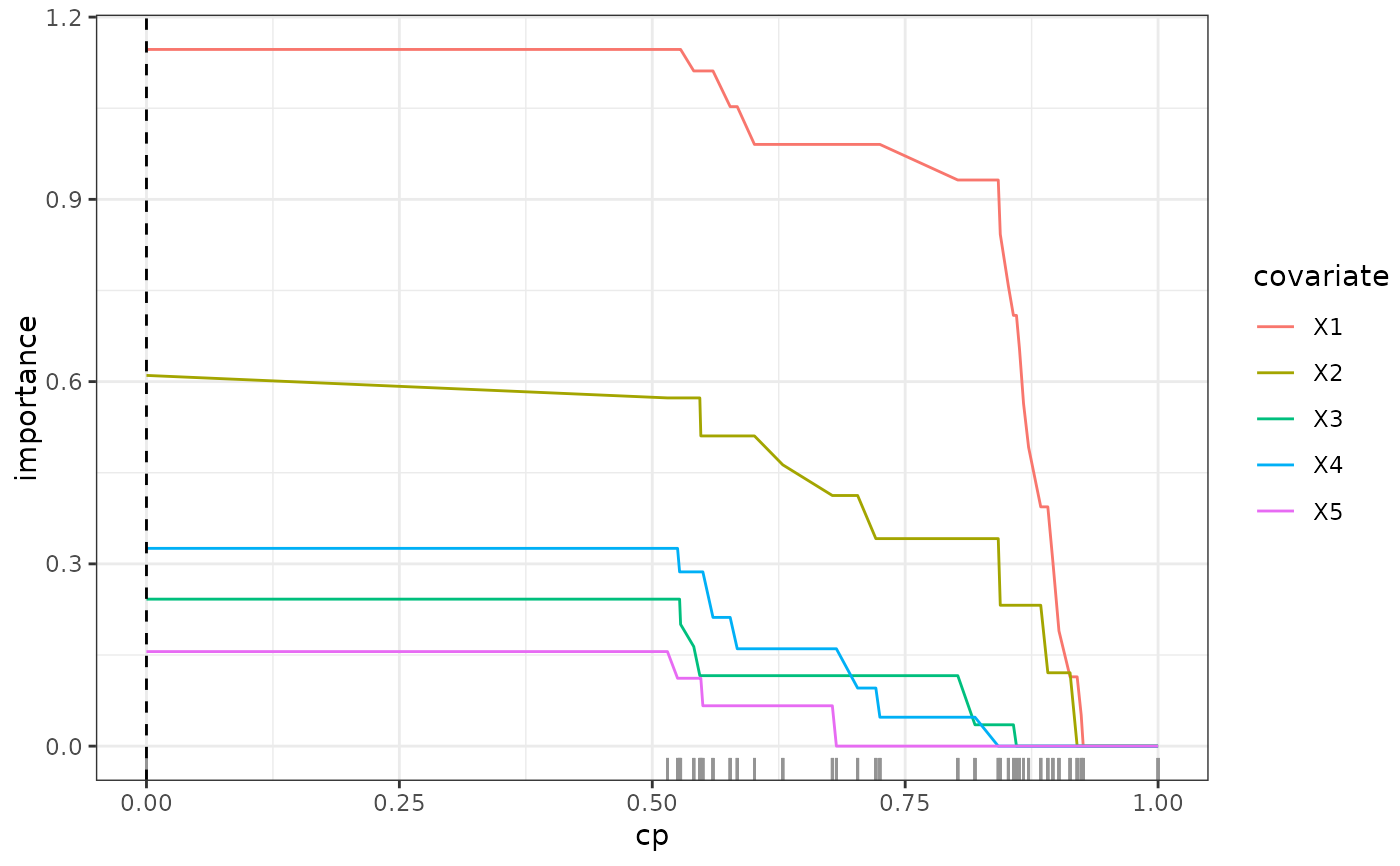
This function calculates the variable importance of an SDForest and the out-of-bag performance for different complexity parameters.
# S3 method for class 'SDForest'
regPath(object, cp_seq = NULL, X = NULL, Y = NULL, Q = NULL, copy = TRUE, ...)Arguments
- object
an SDForest object
- cp_seq
A sequence of complexity parameters. If NULL, the sequence is calculated automatically using only relevant values.
- X
The training data, if NULL the data from the forest object is used.
- Y
The training response variable, if NULL the data from the forest object is used.
- Q
The transformation matrix, if NULL the data from the forest object is used.
- copy
Whether the tree should be copied for the regularization path. If FALSE, the pruning is done in place and will change the SDForest. This might be reasonable, if the SDForest is to large to copy.
- ...
Further arguments passed to or from other methods.
Value
An object of class paths containing
- cp
The sequence of complexity parameters.
- varImp_path
A
matrixwith the variable importance for each complexity parameter.- loss_path
A
matrixwith the out-of-bag performance for each complexity parameter.- cp_min
The complexity parameter with the lowest out-of-bag performance.
- type
Path type