Estimates a regression tree using spectral deconfounding. A regression tree is part of the function class of step functions \(f(X) = \sum_{m = 1}^M 1_{\{X \in R_m\}} c_m\), where (\(R_m\)) with \(m = 1, \ldots, M\) are regions dividing the space of \(\mathbb{R}^p\) into \(M\) rectangular parts. Each region has response level \(c_m \in \mathbb{R}\). For the training data, we can write the step function as \(f(\mathbf{X}) = \mathcal{P} c\) where \(\mathcal{P} \in \{0, 1\}^{n \times M}\) is an indicator matrix encoding to which region an observation belongs and \(c \in \mathbb{R}^M\) is a vector containing the levels corresponding to the different regions. This function then minimizes $$(\hat{\mathcal{P}}, \hat{c}) = \text{argmin}_{\mathcal{P}' \in \{0, 1\}^{n \times M}, c' \in \mathbb{R}^ {M}} \frac{||Q(\mathbf{Y} - \mathcal{P'} c')||_2^2}{n}$$ We find \(\hat{\mathcal{P}}\) by using the tree structure and repeated splitting of the leaves, similar to the original cart algorithm Breiman2017ClassificationTreesSDModels. Since comparing all possibilities for \(\mathcal{P}\) is impossible, we let a tree grow greedily. Given the current tree, we iterate over all leaves and all possible splits. We choose the one that reduces the spectral loss the most and estimate after each split all the leave estimates \(\hat{c} = \text{argmin}_{c' \in \mathbb{R}^M} \frac{||Q\mathbf{Y} - Q\mathcal{P} c'||_2^2}{n}\) which is just a linear regression problem. This is repeated until the loss decreases less than a minimum loss decrease after a split. The minimum loss decrease equals a cost-complexity parameter \(cp\) times the initial loss when only an overall mean is estimated. The cost-complexity parameter \(cp\) controls the complexity of a regression tree and acts as a regularization parameter.
SDTree(
formula = NULL,
data = NULL,
x = NULL,
y = NULL,
max_leaves = NULL,
cp = 0.01,
min_sample = 5,
mtry = NULL,
fast = TRUE,
Q_type = "trim",
trim_quantile = 0.5,
q_hat = 0,
Qf = NULL,
A = NULL,
gamma = 0.5,
gpu = FALSE,
mem_size = 1e+07,
max_candidates = 100,
Q_scale = TRUE
)Arguments
- formula
Object of class
formulaor describing the model to fit of the formy ~ x1 + x2 + ...whereyis a numeric response andx1, x2, ...are vectors of covariates. Interactions are not supported.- data
Training data of class
data.framecontaining the variables in the model.- x
Matrix of covariates, alternative to
formulaanddata.- y
Vector of responses, alternative to
formulaanddata.- max_leaves
Maximum number of leaves for the grown tree.
- cp
Complexity parameter, minimum loss decrease to split a node. A split is only performed if the loss decrease is larger than
cp * initial_loss, whereinitial_lossis the loss of the initial estimate using only a stump.- min_sample
Minimum number of observations per leaf. A split is only performed if both resulting leaves have at least
min_sampleobservations.- mtry
Number of randomly selected covariates to consider for a split, if
NULLall covariates are available for each split.- fast
If
TRUE, only the optimal splits in the new leaves are evaluated and the previously optimal splits and their potential loss-decrease are reused. IfFALSEall possible splits in all the leaves are reevaluated after every split.- Q_type
Type of deconfounding, one of 'trim', 'pca', 'no_deconfounding'. 'trim' corresponds to the Trim transform Cevid2020SpectralModelsSDModels as implemented in the Doubly debiased lasso Guo2022DoublyConfoundingSDModels, 'pca' to the PCA transformationPaul2008PreconditioningProblemsSDModels. See
get_Q.- trim_quantile
Quantile for Trim transform, only needed for trim, see
get_Q.- q_hat
Assumed confounding dimension, only needed for pca, see
get_Q.- Qf
Spectral transformation, if
NULLit is internally estimated usingget_Q.- A
Numerical Anchor of class
matrix. Seeget_W.- gamma
Strength of distributional robustness, \(\gamma \in [0, \infty]\). See
get_W.- gpu
If
TRUE, the calculations are performed on the GPU. If it is properly set up.- mem_size
Amount of split candidates that can be evaluated at once. This is a trade-off between memory and speed can be decreased if either the memory is not sufficient or the gpu is to small.
- max_candidates
Maximum number of split points that are proposed at each node for each covariate.
- Q_scale
Should data be scaled to estimate the spectral transformation? Default is
TRUEto not reduce the signal of high variance covariates, and we do not know of a scenario where this hurts.
Value
Object of class SDTree containing
- predictions
Predictions for the training set.
- tree
The estimated tree of class
Nodefrom Glur2023Data.tree:StructureSDModels. The tree contains the information about all the splits and the resulting estimates.- var_names
Names of the covariates in the training data.
- var_importance
Variable importance of the covariates. The variable importance is calculated as the sum of the decrease in the loss function resulting from all splits that use this covariate.
References
Examples
set.seed(1)
n <- 10
X <- matrix(rnorm(n * 5), nrow = n)
y <- sign(X[, 1]) * 3 + rnorm(n)
model <- SDTree(x = X, y = y, cp = 0.5)
# \donttest{
set.seed(42)
# simulation of confounded data
sim_data <- simulate_data_step(q = 2, p = 15, n = 100, m = 2)
X <- sim_data$X
Y <- sim_data$Y
train_data <- data.frame(X, Y)
# causal parents of y
sim_data$j
#> [1] 4 15
tree_plain_cv <- cvSDTree(Y ~ ., train_data, Q_type = "no_deconfounding")
tree_plain <- SDTree(Y ~ ., train_data, Q_type = "no_deconfounding", cp = 0)
tree_causal_cv <- cvSDTree(Y ~ ., train_data)
tree_causal <- SDTree(y = Y, x = X, cp = 0)
# check regularization path of variable importance
path <- regPath(tree_causal)
plot(path)
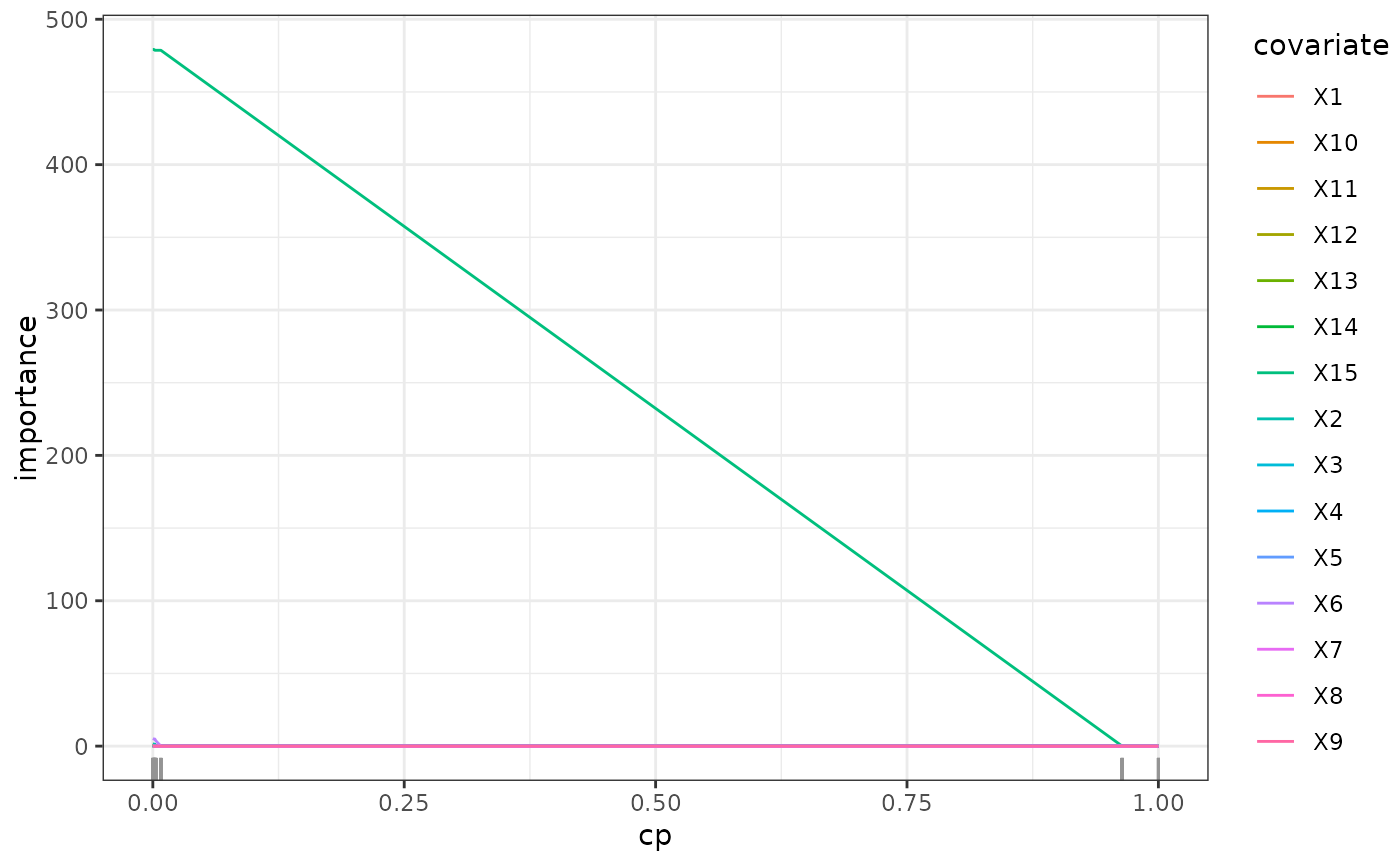 tree_plain <- prune(tree_plain, cp = tree_plain_cv$cp_min)
tree_causal <- prune(tree_causal, cp = tree_causal_cv$cp_min)
plot(tree_causal)
plot(tree_plain)
# }
tree_plain <- prune(tree_plain, cp = tree_plain_cv$cp_min)
tree_causal <- prune(tree_causal, cp = tree_causal_cv$cp_min)
plot(tree_causal)
plot(tree_plain)
# }